Critical Threat Detected: Genomic Analysis of Klebsiella pneumoniae ST17 Co-producing KPC-2 and NDM-1
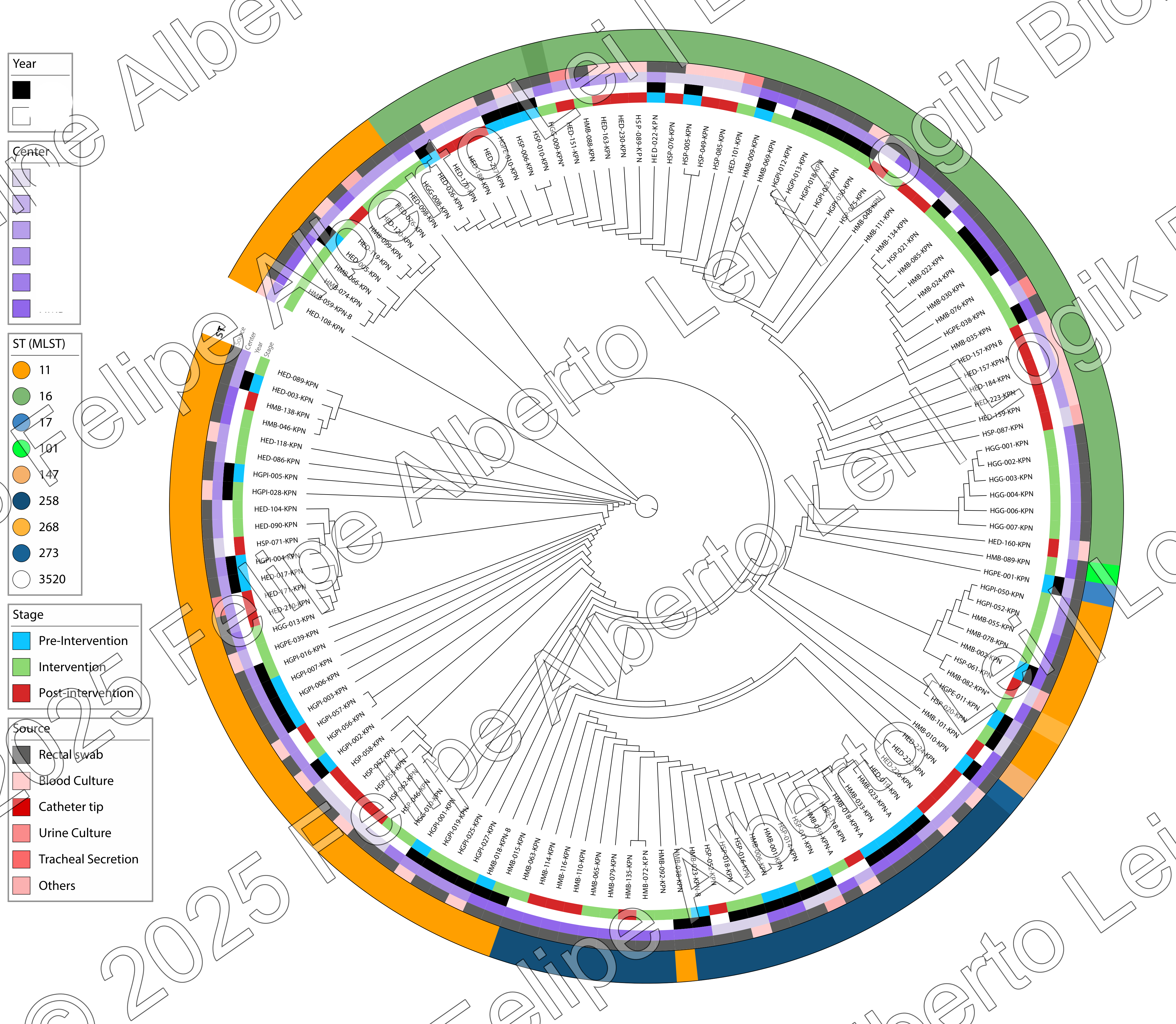
The genomic analysis of the HGPE-001-KPN isolate reveals an adversary of maximum risk. This pathogen belongs to the rare ST17 clone and carries a genetic arsenal that makes it resistant to almost all available treatment options, representing the pinnacle of antimicrobial resistance evolution. This article presents a visual and technical analysis of the genomic data from this critical isolate.
Profile of a Hyper-Resistant Pathogen
The genomic analysis of the HGPE-001-KPN isolate reveals multiple resistance mechanisms highly concentrated in a single microorganism. This isolate belongs to the rare ST17 clone and carries a genetic arsenal that makes it resistant to almost all available treatment options.
Sequence Type
Rare clone, but of the highest risk due to extreme resistance profile.
Dual Threat
Co-production of carbapenemases
Resistance Genes
The highest number in the entire studied cohort.
The Complete Resistome: An Incomparable Genetic Arsenal
The ST17 isolate possesses the highest number of resistance genes (18 in total) in the entire studied cohort. This graph illustrates the diversity of mechanisms that confer resistance to multiple antibiotic classes, explaining its hyper-resistance phenotype.
The Therapeutic Impasse of Co-Production
The coexistence of KPC-2 and NDM-1 in a single pathogen neutralizes the main treatment strategies for infections by multidrug-resistant bacteria, creating a scenario of cascading therapeutic failure.
| Last-Generation Antibiotic | Action against KPC-2 | Action against NDM-1 | Result in Co-Producer |
|---|---|---|---|
| Ceftazidime-Avibactam | ✓ | ✗ | ✗ FAILURE |
| Aztreonam | ✗ | ✓ | ✗ FAILURE |
Avibactam inhibits KPC but not NDM. Aztreonam is not affected by NDM but is destroyed by KPC. The result is resistance to both therapeutic strategies.
Contextualizing the Threat
Although ST17 is rare (only 1 isolate in a cohort of 176), its existence is a warning signal of continuous resistance evolution. It emerges from a scenario dominated by other high-risk clones, such as ST11.
Clone Distribution in Cohort
The Ecosystem of Resistance
ST11: The Dominant Clone
Representing 35.8% of the cohort, ST11-KPC is the endemic "reservoir," establishing the foundation for resistance in the region.
Plasmids: Vectors of Resistance
Genes like
bla
NDM-1
circulate in mobile plasmids, transferred to already established clones.
ST17: The Point of Convergence
Represents the result of this convergence, where multiple resistance mechanisms unite, creating a superbacteria.
Call to Action: Surveillance and Control
Detection of an isolate like HGPE-001-KPN requires an immediate and coordinated response, focused on containment, rapid diagnosis, and genomic surveillance.
Genomic Surveillance
Implement routine sequencing to rapidly identify high-risk clones.
Maximum Containment
Strict contact isolation and epidemiological investigation are mandatory.
One Health Approach
Monitor wastewater to detect presence of these pathogens.
Conclusion
This visual analysis of the genomic data from the HGPE-001-KPN isolate and its epidemiological contextualization demonstrates the growing complexity of antimicrobial resistance. The co-production of KPC-2 and NDM-1, combined with the genetic arsenal of ST17, represents one of the most challenging cases in terms of therapeutic options. Integrated surveillance and preventive action are essential to prevent the spread of such highly resistant clones.